High-sensitivity blood testing increases the detection rate of myocardial injury
Published Mar. 06, 2024
By Hopkins Medtech
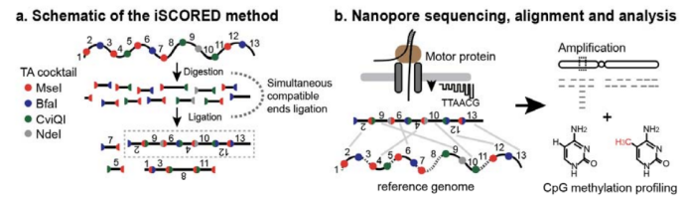
Copy number variations (CNVs) are almost ubiquitous in cancer. In many cases,somatic CNV analysis can identify oncogenic pathways and provide potential therapeutic targets. Recently, a research team from Dartmouth-Hitchcock Medical Center (DHMC) developed a method called iSCORED, which is a one-step random genome DNA reconstruction method capable of efficient and unbiased quantification of CNVs using real-time nanopore sequencing.
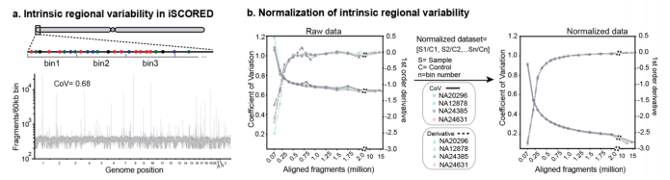
By leveraging long-linked reads, the research team generated approximately 1-2 million genome fragments within one hour of MinION sequencing, allowing for a high-resolution genome dosage comparison. In a cohort of 26 malignant brain tumors,iSCORED demonstrated 100% consistency in CNV detection compared to clinically validated next-generation sequencing and chromosomal microarray results, including chromosomal alterations and amplifications of oncogenes. Additionally,iSCORED enables concurrent methylation classification of brain tumors without the need for additional tissue preparation. Integrated methylation information revealed low methylation in the promoters of all detected amplified oncogenes. The entire workflow, including the automatic generation of CNV and methylation reports, showcases the potential of iSCORED in advancing efficient and comprehensive genomic analyses for cancer research.
CNV activates oncogenes and inactivates tumor suppressor genes, driving the occurrence and progression of cancer. These effects make copy number analysis an integral component of tumor grading and diagnosis. However, the current dependence on nucleic acid hybridization and next-generation sequencing means that CNV analysis is confined to highly complex centralized laboratories and typically takes several days at a minimum.
A faster, cheaper, and simpler method for copy number variation (CNV) analysis can enhance clinical decision-making, optimize surgical planning, and enable the identification of potential molecular therapies during surgery. Researchers at DHMC found that nanopore sequencing is an improved approach for CNV analysis.
The team described the use of Oxford Nanopore Technologies’ MinION device in CNV analysis. This device provides real-time resolution of long-read nucleotide sequences. To apply this technology to CNV detection, researchers randomly analyzed connected DNA fragments. This method can identify multiple locatable DNA segments in a single sequencing read. Additionally, the cost per sample is very low, at only $125, and it is easy to establish a sequencing platform, with the budget for MinION being $6,000-8,000 and the device budget for PromethION being $14,000-16,000, making it an economical choice for clinical applications.
The DHMC team studied 26 malignant brain tumors, and the nanopore analysis detected changes and amplifications identical to clinically validated next-generation sequencing and chromosomal microarray results. The method allows simultaneous tumor methylation classification without additional tissue preparation. Low methylation in the promoters of all detected amplified oncogenes was found. Currently, the CNV analysis method is undergoing patent application and is called “irreversible Sticking Compatible Overhang to Reconstruct DNA (iSCORED).” Researchers view accelerating CNV analysis as a way to shorten the time to identify patients suitable for molecular-targeted drug therapy.

