A PCR Method for Subtyping Small Cell Lung Cancer Developed by MD Anderson Cancer Center
Small Cell Lung Cancer (SCLC), a highly aggressive variant of lung cancer, constitutes about 15% of all lung cancer diagnoses. Geographic variations in SCLC incidence are noted, with a decreasing trend observed yearly, attributed to enhanced public health initiatives and a decline in smoking rates. Nonetheless, SCLC continues to be particularly detrimental due to its fast proliferation and tendency to spread quickly. Although subtyping in clinical contexts can improve treatment outcomes, obtaining sufficient tissue for accurate pathological subtyping remains a challenge. In response to this, researchers have recently innovated a sensitive PCR technique for more precise subtyping of SCLC.

By delving into the intricate epigenetic mechanisms that regulate the transcriptional landscape of SCLC, scientists proposed that detecting unique methylation patterns in either tumor tissue or the blood of SCLC patients could delineate specific subtypes. In a pioneering effort, Nucleix, based in San Diego, California, employed their EpiCheck platform to introduce a methylation-focused PCR detection approach, distinguishing SCLC subtypes. This method merges the use of methylation-sensitive restriction enzyme (MSRE) processing with quantitative PCR (qPCR) to pinpoint methylation variances at the genomic scale.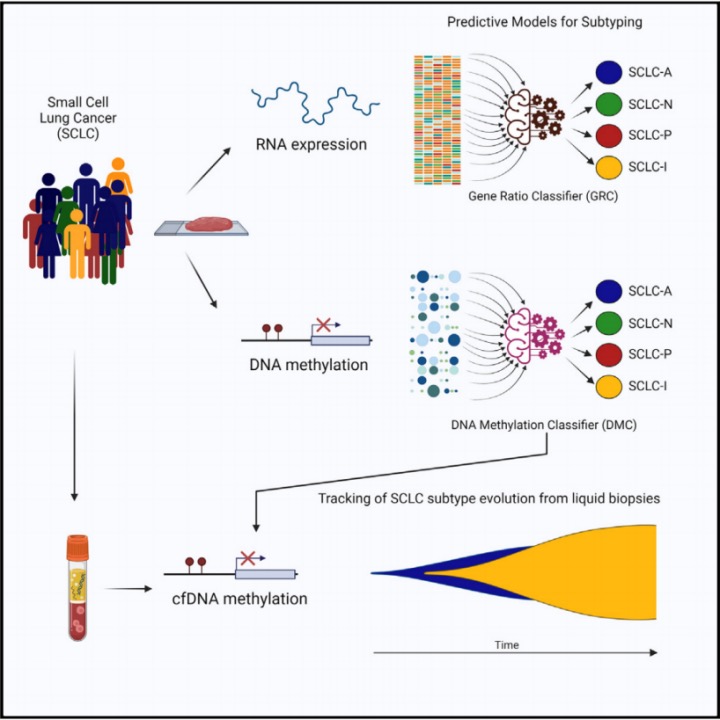
Leveraging prior success in identifying SCLC through plasma samples from heavy smokers based on DNA methylation—with a remarkable sensitivity of 94% and specificity of 95%—Nucleix unveiled a 13-marker PCR detection strategy. This development not only expedites the interval from diagnosis to specialized treatment plans but also introduces novel biomarkers for classifying SCLC into subtypes. Impressively, in a blind study, this 13-marker PCR accomplished a 97% accuracy rate in classifying SCLC tissue samples.
Engaging with two separate groups consisting of 179 SCLC patients, the research team utilized whole-genome bisulfite sequencing (RRBS) along with machine learning techniques to craft a reliable DNA methylation-based classifier (SCLC-DMC) capable of discriminating between SCLC subtypes. Further optimization of this classifier to detect circulating-free DNA (cfDNA) demonstrated its potential to differentiate SCLC subtypes in plasma, revealing the dynamic nature of SCLC phenotypes over the course of the disease and underscoring the value of continuous monitoring in clinical management. These findings indicate that methylation profiles, from both tumor and cfDNA samples, could be instrumental in identifying SCLC subtypes, thereby informing targeted treatment strategies.
In conclusion, the advent of this sophisticated PCR method for delineating SCLC subtypes marks a pivotal leap in cancer diagnostics. Through the integration of methylation-based markers and machine learning, this methodology is set to enhance the precision and efficiency of SCLC diagnosis and treatment, offering patients more personalized therapeutic avenues. As advancements in research press forward, the promise for such technologies to revolutionize cancer care continues to be significant and compelling.
For additional insights on this technological breakthrough and its impact on SCLC management, stay connected with Nucleix for forthcoming updates. Keep abreast of the latest developments in lung cancer diagnostics and treatment methods. United in our efforts, we can advance in the battle against cancer.